Difference between revisions of "XBio:D Javascript Library Reference"
(→showTaxonHierarchy) |
(→Options) |
||
| Line 212: | Line 212: | ||
* options | * options | ||
===== Options ===== | ===== Options ===== | ||
| − | * widget_options - | + | * widget_options - Object |
==== showOccurrenceInfo ==== | ==== showOccurrenceInfo ==== | ||
Revision as of 21:17, 20 April 2015
Introduction
The xBio:D JavaScript library offers the functionality to create rich, dynamic, and interactive features for use with the OJ_Break API. The OJ_Break API provides procedures to retrieve data within the xBio:D database, and the xBio:D JS library is how that data gets presented. Users planning on working with the xBio:D JS library should be familiar with JavaScript programming and object-oriented programming.
Contents
- 1 API Access
- 2 Example
- 3 Reference
- 3.1 Agent
- 3.2 Occurrence
- 3.3 Search
- 3.4 Taxon
- 3.5 Visual
API Access
All functionality in the xBio:D JS library depends on access of data from the OJ_Break API. See OJ_Break API Access for an overview of the API and how to obtain an API access key.
Example
An easy way to become familiar with the xBio:D JS library is to see a simple example of a web application which uses two xBio:D methods. The first is a Google Maps widget which takes data from the OJ_Break method getLocalities for a specified tnuid and displays the data onto an interactive map. Read more about the Google Maps JavaScript API. The second method used in the example is a listing of included taxa for a specified tnuid by using the OJ_Break method getIncludedTaxa.
1 <!DOCTYPE HTML> 2 <html> 3 <head> 4 <meta charset="UTF-8"/> 5 <title>xBio:D API Test</title> 6 <link rel="stylesheet" type="text/css" href="xbiod.css"> 7 <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.8/jquery.min.js"></script> 8 <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jqueryui/1.10.4/jquery-ui.min.js"></script> 9 <script type="text/javascript" src="https://maps.googleapis.com/maps/api/js?sensor=false"></script> 10 <script type="text/javascript" src="http://osuc.biosci.ohio-state.edu/JSLib/xbiod_lib/xbiod.js"></script> 11 <script> 12 // Call the loader for the current page 13 $(document).ready(loader); 14 15 function loader() { 16 17 // Initialize xBio:D library 18 xbiod.init(['visual','taxon'], /*API_KEY*/, function() { 19 var tnuid = 605; 20 21 // Load xBio:D components 22 var visual = new xbiod.visual(); 23 var taxon = new xbiod.taxon(); 24 25 // Load map 26 visual.showGoogleMap('map_id', tnuid); 27 28 // Load included taxa 29 taxon.showIncludedTaxa('included_id', tnuid, {show_num_spms: 'Y'}); 31 32 }); 33 } 34 </script> 35 <style type="text/css"> 36 html { height: 100% } 37 body { height: 100%; margin: 0; padding: 0 } 38 #map_id { height: 50% } 39 #included_id { height: 100% } 40 </style> 41 </head> 42 <body> 43 <div id="map_id"/> 44 <div id="included_id"/> 45 </body> 46 </html>
There are a few things to note about this example:
- The <!DOCTYPE html> tag is necessary for HTML 5 applications (line 1)
- Jquery 1.8 library is included with a <script> tag (line 7)
- Jquery UI 1.10.4 library is included with a <script> tag (line 8)
- The Google Map API JavaScript library is included with a <script> tag (line 9)
- The xBio:D JS library is included with a <script> tag (line 10)
- A function called loader intializes the xBio:D resources when the web page has finished loading (line 15)
- Two objects where created to access the xBio:D components called visual and taxon (line 22 - 23)
- The applications are created by calling there respective methods via the xBio:D library components (line 26 & 29 - 30)
- In the <body> there are two <div> elements which create areas to hold the widgets. The first is called map_id and the second is included_id. (line 43 - 44)
These steps will be explained below.
Declaring Application as HTML 5
It is recommended for any web application to be declared as a true DOCTYPE. This can easily be done by using the HTML 5 DOCTYPE as seen below. This allows the application to be more cross-browser compliant. Please refer to the Google Maps Documentation on HTML 5 for more info.
<!DOCTYPE html>
Loading JQuery Library
<script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.8/jquery.min.js"></script>
The URL in the <script> tag above is the location of the JavaScript file which allows JQuery 1.8 to run on the web application. JQuery is used in the xBio:D JS library to get methods from the OJ_Break API and other scripts. It is a necessary component of any web app that intends to use xBio:D functionality.
Loading the Google Maps API
<script type="text/javascript" src="https://maps.googleapis.com/maps/api/js?sensor=false"></script>
The URL in the <script> tag above is the location of the JavaScript file which will load all the necessary defintions to allow access to the Maps API. Normally, an access key would need to be provided in order to use the Maps library. See Loading the Maps API for more info. This script file is necessary for web apps that use Google Maps as part of the application. It is a necessary file to have in this simple example, but is not required for all xBio:D applications.
Loading the xBio:D JavaScript Library
<script type="text/javascript" src="http://osuc.biosci.ohio-state.edu/JSLib/xbiod_lib/xbiod.js"></script>
The URL in the <script> tage above is the location of the xBio:D JavaScript Library which is a required file for this simple example and any web application that will be using xBio:D applications.
Initializing the xBio:D JS Library
<script> // Call the loader for the current page $(document).ready(loader);
function loader() {
// Initialize xBio:D library xbiod.init(['visual','taxon'], /*API_KEY*/, function(){
To begin using the xBio:D library, first the components need to be loaded. This is done through the xBio:D init function which takes three parameters: an array of the components to be loaded, an API access key, and a callback function. In this example, the visual and taxon resources are passed to the init function for loading (all resources and their methods can be found in the Reference section below). This example also shows where a user would provide an OJ_Break API access key. See OJ_Break API Access for more information on access keys. Lastly, a callback method needs to be provided which will create the component objects.
Component Objects
// Load xBio:D components var visual = new xbiod.visual(); var taxon = new xbiod.taxon();
An object derived from a namespace is instatiated using the new keyword and a namespace's constructor. The JavaScript namespace that represents the xBio:D visual applications is the visual namespace/component and similarly the namespace that represents the xBio:D taxon applications is the taxon namespace/component. Access to functionality from either of these components requires an object with a reference to that component. These objects allow methods from each component to be called to the web page.
Calling Application Methods
// Load map visual.showGoogleMap('map_id', tnuid); // Load included taxa taxon.showIncludedTaxa('included_id', tnuid, {show_num_spms: 'Y', taxonFormat: '<a href="test.html?tnuid=%tnuid%">%taxon%</a> %author% - %num_spms%'});
Each application has a method associated with it which takes several parameters which will dictate what data it loads, where it will be contained, and options for how it should be presented. These methods and requirements can be found in the Reference section.
In the above example, the visual method showGoogleMap gets passed a <div> element ID string which decides where it will be contained and a tnuid which will dictate which taxon data the map will locate.
The taxon method showIncludedTaxa gets passed similar parameters (a specified <div> element ID string and a tnuid) however it also gets additional options in the form of a JavaScript object literal. In this example, the option show_num_spms is given the Boolean_flag 'Y' signifying that the application should display the number of specimens for that particular taxon and taxonFormat is assigned to present the data as links using the <a> tag.
Element Containers
Each method requires one or more unique <div> tag IDs as string parameters (seen in the example <body> below). This will be used to specify where the resource will be contained in the web page.
<body> <div id="map_id"/> <div id="included_id"/> </body>
Reference
Agent
showAgentInfo
Description
Displays information about an agent, including name, contact information, and a short biography.
Parameters
Options
- widget_options - Object
showAgentOccurrences
Description
Displays collected occurrences for a specified agent on a Google Map. Creates two windows.
Parameters
Options
- info_element_id - ID String (defaults to element_id parameter)
- generalFormat - HTML String
- markers - String ('spiderfy' or 'cluster')
- offset - Number
- limit - Number
- icons - Object
- widget_options
Additional Requirements
showAgentOccurrenceLocalityInfo
Description
Displays locality information for a certain agent's collecting occurrence.
Parameters
Options
None
showDescribedTaxa
Description
Displays all taxa described by the specified agent.
Parameters
Options
- taxonFormat - String
- useTaxonItalics - Boolean
- limit - Number
- offset - Number
- widget_options - Object
showPublications
Description
Displays all publications for a specified agent.
Parameters
Options
- limit - Number
- offset - Number
- widget_options - Object
Occurrence
showBiologicalInfo
Description
Displays both the habitat and associations for a selected occurrence identifier (cuid).
Parameters
Options
- widget_options - Object
showOccurrenceInfo
Description
Displays collecting trip info for a specified occurrence.
Parameters
Options
- widget_options - Object
showDeterminations
Description
Displays determinations for a specified occurence.
Parameters
Options
- widget_options - Object
showLocalityInfo
Description
Displays locality info for a specified occurrence.
Parameters
Options
- widget_options - Object
showSpecimenInfo
Description
Displays specimen info for a specified occurrence.
Parameters
Options
- widget_options - Object
Search
showSearch
Description
A JQuery UI autocomplete search bar which can search for taxa, cuids, institutions, agents, journals, organizations, collecting methods, localities, and places.
Parameters
- element_id
- options
Options
- search - the type of search to execute. Controls the passing of URL parameters to methods. Can be specified a either 'id' or 'name'. Defaults to 'id'
- category - array of the below object elements
- type - a category keyword that will control which data gets searched for
- handler_url - a URL that will be passed the ID of the selected type option
Additional Information
Category Options Formatting
Below is an example of how the categories should be input:
// Load search search.showSearch('search_id', {category: [{type: 'taxa', handler_url: 'http://hol.osu.edu/index.html'}, {type: 'cuids', handler_url: 'http://hol.osu.edu/spmInfo.html'}]});
Valid Category Keywords
Below is a list of available category keywords that the category array option can take (Note: the keyword spelling must match exactly what is seen below):
- all
- taxa
- cuids
- inst
- agents
- journals
- orgs
- coll_methods
- locs
- places
Taxon
showAssociations
Description
Displays all associations for a specified taxon.
Parameters
- element_id
- tnuid
- options
Options
- basic_only - Boolean_flag
- limit - Number
- offset - Number
- widget_options
showInstitutions
Description
Displays institutions for which specimens of the selected taxon level belong.
Parameters
- element_id
- tnuid
- options
Options
- limit - Number
- offset - Number
- widget_options
showContributors
Description
Displays all contributing agents related to the occurrence, taxonomy, literature, and media of a taxon.
Parameters
- element_id
- tnuid
- options
Options
- inst_id
- widget_options
showDeterminers
Description
Displays determiners for a specified taxon.
Parameters
- element_id
- tnuid
- options
Options
- limit - Number
- offset - Number
- inst_id
- widget_options
showHabitats
Description
Displays all identified habitats for a selected taxon.
Parameters
- element_id
- tnuid
- options
Options
- limit - Number
- offset - Number
- widget_options
showIncludedTaxa
Description
Displays taxa directly included within a taxon but not synonymous with it.
Parameters
- element_id
- tnuid
- options
Options
- inst_id
- show_syns
- show_fossiles
- types_only
- useTaxonItalics - Boolean
- limit - Number
- offset - Number
- widget_options
showLiterature
Description
Displays relevent publications for a specified taxon with links to the literature when available.
Parameters
- element_id
- tnuid
- options
Options
- show_children - Boolean_flag
- show_bib - Boolean_flag
- useTaxonItalics - Boolean
- limit - Number
- offset - Number
- widget_options
showSubordinateTaxa
Description
Displays the count of valid taxa levels available below the current taxon rank.
Parameters
- element_id
- tnuid
- options
Options
- inst_id
- widget_options
showSynonyms
Description
Displays all taxa which are objectively or subjectively synonymous with a selected taxon.
Parameters
- element_id
- tnuid
- options
Options
- show_fossils
- useTaxonItalics - boolean
- limit - Number
- offset - Number
- widget_options
showTaxonMedia
Description
Displays a table of all media associated to a specific taxon rank.
Parameters
- element_id
- tnuid
- options
Options
- inst_id
- row_count - Number
- media_type
- limit - Number
- offset - Number
- widget_options
showTypes
Description
Displays information and specimen types for a specified taxon.
Parameters
- element_id
- tnuid
- options
Options
- show_children
- inst_id
- primary_only - Boolean_flag
- basic_only - Boolean_flag
- offset - number
- limit - number
- useTaxonItalics - boolean
- widget_options
Visual
showGoogleMap
Description
A Google map which displays locality markers for a specified taxon.
Parameters
Options
Additional Requirements
showTaxonHierarchy
Description
An interactive, tree-based animation which allows a user to navigate through a selected taxon's hierarchy and all taxa available through the OJ_Break API.
Parameters
Options
- show_num_spms
- inst_id
- show_syns
- show_fossils
- types_only
- nodeColor - string or hexadecimal number
- backgroundColor - string or hexadecimal number
- handler_url - URL string
- size - JavaScript object with two string properties:
- width - either a number or string in pixels
- height - either a number of string in pixels
- imagePath - string of relative path to a folder containing necessary images.
- widget_options
Additional Requirements
- Processing.js 1.4.1+
- Pass requestedNode parameter a value of null when calling function. See below:
// Load taxon hierarchy visual.showTaxonHierarchy('canvas_id', tnuid, null, { /*options*/ });
Additional Information
Valid Color Options
Below is a list of the available colors that can be used as the background or node color:
- red
- scarlet
- orange
- yellow
- green
- blue
- cyan
- violet
- purple
- grey/gray
- white
NOTE: any color (that is, not limited to the above list) can be chosen by specifying its hexadecimal color string (for example, '#70CFF5').
URL Handler Option
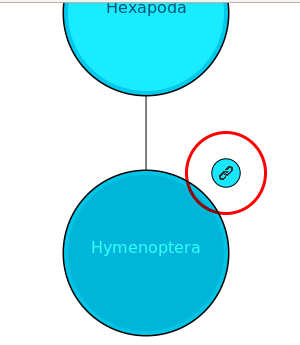
The handler_url option can be used to allow the nodes to have links attached to them, so that a user may follow the link for more information on that specific taxon.
// Load taxon hierarchy visual.showTaxonHierarchy('canvas_id', tnuid, null, {nodeColor: 'cyan', backgroundColor: '#FFFFFF', handler_url: 'http://hol.osu.edu/index.html'});
In this example, a default web page URL is given (note the use of http:// at the beginning). When provided a URL, the taxon hierachy animation will present a link button to the top right of the node that is currently being highlighted (see image below). This link will not be present if a URL is not specified. If this button is clicked, the handler_url link will be followed.
Size Option
The taxon hierarchy application can be resized in one of two ways: via CSS (preferred) or by inputting the dimensions of the HTML canvas into the API method call as options. One way this can be done is by specifying the width and height as numbers:
// Load taxon hierarchy visual.showTaxonHierarchy('canvas_id', tnuid, null, {size: {width: 300, height: 900}});
The dimensions of the canvas can also be input as pixels:
// Load taxon hierarchy visual.showTaxonHierarchy('canvas_id', tnuid, null, {size: {width: "300px", height: "900px"}});
Both of these methods (CSS and parameter passing) are valid, but they should never both be used at the same time since unexpected errors may occur.
Image Path Option
Due to security restrictions, Processingjs is limited in its function to request images. As a consequence, any user wishing to use the animation must have copies of the required images available locally on their server. To access these images, a relative directory path must be input in the form of a optional parameter. The call may look something like this:
// Load taxon hierarchy visual.showTaxonHierarchy('canvas_id', tnuid, null, {imagePath: "media/images/"});
Portions of this page are modifications based on work created and shared by Google and used according to terms described in the Creative Commons 3.0 Attribution License.