Difference between revisions of "OJ Break v2 Taxon Reference"
(→TaxonReturnIncluded (extends TaxonReturn)) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
'''Introduction''' | '''Introduction''' | ||
| − | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with | + | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication. |
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | ||
| Line 13: | Line 13: | ||
Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | ||
| + | |||
| + | |||
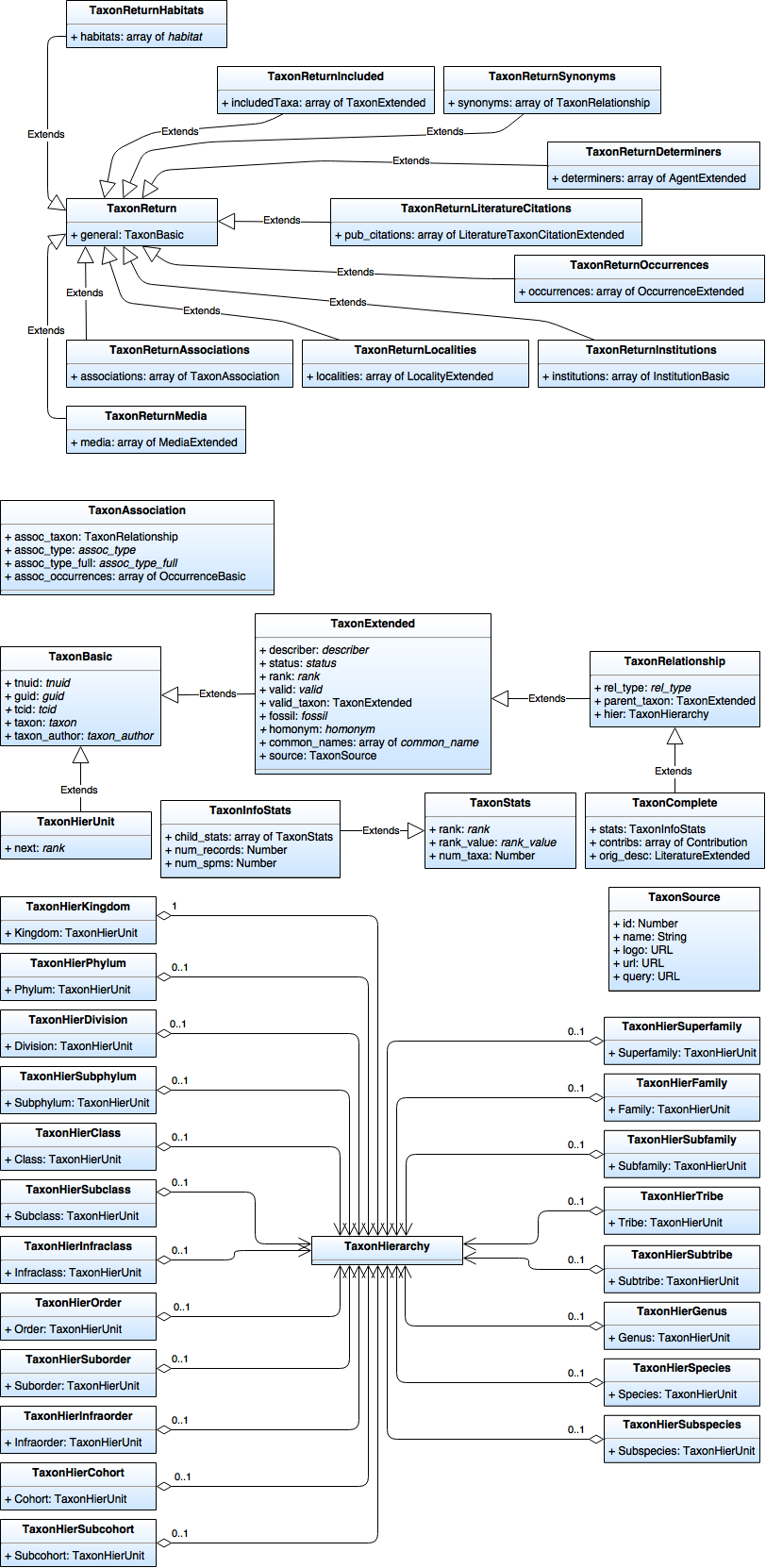
| + | [[File:OJ_Break Data Model - Taxon.png|none|frame|Taxon Data Model]] | ||
== Procedural Reference == | == Procedural Reference == | ||
| Line 241: | Line 244: | ||
==== TaxonBasic ==== | ==== TaxonBasic ==== | ||
* ''tnuid'': ''[[#tnuid|tnuid]]'' | * ''tnuid'': ''[[#tnuid|tnuid]]'' | ||
| + | * ''guid'': ''[[#guid|guid]]'' | ||
| + | * ''tcid'': ''[[#tcid|tcid]]'' | ||
* ''taxon'': ''[[#taxon|taxon]]'' | * ''taxon'': ''[[#taxon|taxon]]'' | ||
* ''taxon_author'': ''[[#taxon_author|taxon_author]]'' | * ''taxon_author'': ''[[#taxon_author|taxon_author]]'' | ||
| Line 353: | Line 358: | ||
==== TaxonRelationship (extends [[#TaxonExtended (extends TaxonBasic)|TaxonExtended]]) ==== | ==== TaxonRelationship (extends [[#TaxonExtended (extends TaxonBasic)|TaxonExtended]]) ==== | ||
* ''rel_type'': ''[[#rel_type|rel_type]]'' | * ''rel_type'': ''[[#rel_type|rel_type]]'' | ||
| − | * ''parent_taxon'': [[# | + | * ''parent_taxon'': [[#TaxonExtended|TaxonBasic]] |
* ''hier'': [[#TaxonHierarchy|TaxonHierarchy]] | * ''hier'': [[#TaxonHierarchy|TaxonHierarchy]] | ||
| Line 369: | Line 374: | ||
==== TaxonReturnIncluded (extends [[#TaxonReturn|TaxonReturn]]) ==== | ==== TaxonReturnIncluded (extends [[#TaxonReturn|TaxonReturn]]) ==== | ||
| − | * ''includedTaxa'': array of [[#TaxonExtended (extends TaxonBasic)| | + | * ''includedTaxa'': array of [[#TaxonExtended (extends TaxonBasic)|TaxonExtended]] |
==== TaxonReturnInstitutions (extends [[#TaxonReturn|TaxonReturn]]) ==== | ==== TaxonReturnInstitutions (extends [[#TaxonReturn|TaxonReturn]]) ==== | ||
| Line 411: | Line 416: | ||
String - the describer(s) of a taxon. | String - the describer(s) of a taxon. | ||
==== fossil ==== | ==== fossil ==== | ||
| − | String - A modified boolean_flag, either Y, N or B, representing whether a taxon name is a extinct fossil, exclusively extant, or present today and in fossils. | + | String - A modified boolean_flag, either Y, N or B, representing whether a taxon name is a extinct fossil, exclusively extant, or present today and in fossils. |
| + | ==== guid ==== | ||
| + | String - A globally unique identifier for an xBio:D domain resource in the form of a stable URI dereferenced by the ''bioguid.osu.edu'' domain. | ||
==== habitat ==== | ==== habitat ==== | ||
String - Description or identifier of the ecological or environmental area that a particular species is known to inhabit. | String - Description or identifier of the ecological or environmental area that a particular species is known to inhabit. | ||
| Line 421: | Line 428: | ||
Number - A number representing the rank of a taxon with higher numbers representing higher positions in the taxonomic hierarchy. | Number - A number representing the rank of a taxon with higher numbers representing higher positions in the taxonomic hierarchy. | ||
==== rel_type ==== | ==== rel_type ==== | ||
| − | String - The taxonomic (hierarchical) relationship between the taxonomic concept indicated by the taxon name and its parent taxonomic concept. Can be Member, Junior synonym, etc. | + | String - The taxonomic (hierarchical) relationship between the taxonomic concept indicated by the taxon name and its parent taxonomic concept. Can be Member, Junior synonym, etc. |
==== status ==== | ==== status ==== | ||
| − | String - The nomenclatural status of a taxon name using a formalized vocabulary. | + | String - The nomenclatural status of a taxon name using a formalized vocabulary. |
==== taxon ==== | ==== taxon ==== | ||
| − | String - A taxon name string. | + | String - A taxon name string. |
==== taxon_author ==== | ==== taxon_author ==== | ||
| − | String - The author(s) of a taxon with parenthesis surrounding the name of the author(s) if necessary. | + | String - The author(s) of a taxon with parenthesis surrounding the name of the author(s) if necessary. |
| + | ==== tcid ==== | ||
| + | Number - The taxonomic concept identifier which uniquely identifies a taxon concept, which may have many name uses, within xBio:D. | ||
==== tnuid ==== | ==== tnuid ==== | ||
| − | Number - The taxon name use identifier which uniquely identifies a taxon. | + | Number - The taxon name use identifier which uniquely identifies a taxon within xBio:D. |
==== valid ==== | ==== valid ==== | ||
String - A ''Valid'' or ''Invalid'' string representing the validity of a taxon name using subjective interpretations if warranted. | String - A ''Valid'' or ''Invalid'' string representing the validity of a taxon name using subjective interpretations if warranted. | ||
Latest revision as of 13:44, 25 September 2015
Introduction
OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication.
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database.
Contents
- 1 API Information and Access
- 2 Procedural Reference
- 2.1 Taxon
- 2.1.1 getTaxonInfo
- 2.1.2 getTaxonHierarchy
- 2.1.3 getTaxonIncludedTaxa
- 2.1.4 getTaxonSynonyms
- 2.1.5 getTaxonAssociations
- 2.1.6 getTaxonLiteratureCitations
- 2.1.7 getTaxonOccurrences
- 2.1.8 getTaxonTypes
- 2.1.9 getTaxonLocalities
- 2.1.10 getTaxonDeterminers
- 2.1.11 getTaxonInstitutions
- 2.1.12 getTaxonHabitats
- 2.1.13 getTaxonMedia
- 2.1 Taxon
- 3 Taxon Data Type Glossary
- 3.1 Classes
- 3.1.1 TaxonAssociation
- 3.1.2 TaxonBasic
- 3.1.3 TaxonComplete (extends TaxonRelationship)
- 3.1.4 TaxonExtended (extends TaxonBasic)
- 3.1.5 TaxonHierarchy
- 3.1.6 TaxonHierClass
- 3.1.7 TaxonHierCohort
- 3.1.8 TaxonHierDivision
- 3.1.9 TaxonHierFamily
- 3.1.10 TaxonHierGenus
- 3.1.11 TaxonHierInfraclass
- 3.1.12 TaxonHierInfraorder
- 3.1.13 TaxonHierKingdom
- 3.1.14 TaxonHierOrder
- 3.1.15 TaxonHierPhylum
- 3.1.16 TaxonHierSpecies
- 3.1.17 TaxonHierSubclass
- 3.1.18 TaxonHierSubcohort
- 3.1.19 TaxonHierSubfamily
- 3.1.20 TaxonHierSuborder
- 3.1.21 TaxonHierSubphylum
- 3.1.22 TaxonHierSubspecies
- 3.1.23 TaxonHierSubtribe
- 3.1.24 TaxonHierSuperfamily
- 3.1.25 TaxonHierTribe
- 3.1.26 TaxonHierUnit (extends TaxonBasic)
- 3.1.27 TaxonInfoStats (extends TaxonStats)
- 3.1.28 TaxonRelationship (extends TaxonExtended)
- 3.1.29 TaxonReturn
- 3.1.30 TaxonReturnAssociations (extends TaxonReturn)
- 3.1.31 TaxonReturnDeterminers (extends TaxonReturn)
- 3.1.32 TaxonReturnHabitats (extends TaxonReturn)
- 3.1.33 TaxonReturnIncluded (extends TaxonReturn)
- 3.1.34 TaxonReturnInstitutions (extends TaxonReturn)
- 3.1.35 TaxonReturnLiteratureCitations (extends TaxonReturn)
- 3.1.36 TaxonReturnLocalities (extends TaxonReturn)
- 3.1.37 TaxonReturnMedia (extends TaxonReturn)
- 3.1.38 TaxonReturnOccurrences (extends TaxonReturn)
- 3.1.39 TaxonReturnSynonyms (extends TaxonReturn)
- 3.1.40 TaxonSource
- 3.1.41 TaxonStats
- 3.2 Elements
- 3.1 Classes
- 4 See also
API Information and Access
This page specifies the methods and data defined by OJ_Break Version 2, more precisely those that are defined by the Taxon data domain. To get information on any of the other data domains defined in OJ_Break Version 2, visit the See also section.
Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return format, an API access key, and a version number.
Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonInfo?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Notice the specification of the version parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the version parameter has a default value of 2 making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to OJ_Break API Access.
Procedural Reference
Taxon
getTaxonInfo
Description
Parameters
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonInfo?tnuid=30148&inst_id=1&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonHierarchy
Description
Parameters
- tnuid: tnuid
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonHierarchy?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonIncludedTaxa
Description
Parameters
- tnuid: tnuid
- *inst_id: inst_id
- *rank: rank
- *offset: Number
- *limit: Number
- *show_syns: Boolean_flag
- *show_fossils: Boolean_flag
- *types_only: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonIncludedTaxa?tnuid=30148&inst_id=1&show_syns=Y&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonSynonyms
Description
Parameters
- tnuid: tnuid
- *offset: Number
- *limit: Number
- *show_fossils: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonSynonyms?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonAssociations
Description
Parameters
- tnuid: tnuid
- *offset: Number
- *limit: Number
- *basic_only: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonAssociations?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonLiteratureCitations
Description
Parameters
- tnuid: tnuid
- *offset: Number
- *limit: Number
- *show_children: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonLiteratureCitations?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonOccurrences
Description
Parameters
- tnuid: tnuid
- *inst_id: inst_id
- *place_id: place_id
- *offset: Number
- *limit: Number
- *show_children: Boolean_flag
- *basic_only: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonOccurrences?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonTypes
Description
Parameters
- tnuid: tnuid
- *inst_id: inst_id
- *offset: Number
- *limit: Number
- *basic_only: Boolean_flag
- *show_children: Boolean_flag
- *primary_only: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonTypes?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonLocalities
Description
Parameters
- tnuid: tnuid
- *inst_id: inst_id
- *place_id: place_id
- *offset: Number
- *limit: Number
- *show_children: Boolean_flag
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonLocalities?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonDeterminers
Description
Parameters
- tnuid: tnuid
- *inst_id: inst_id
- *offset: Number
- *limit: Number
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonDeterminers?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonInstitutions
Description
Parameters
- tnuid: tnuid
- *offset: Number
- *limit: Number
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonInstitutions?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonHabitats
Description
Parameters
- tnuid: tnuid
- *offset: Number
- *limit: Number
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonHabitats?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getTaxonMedia
Description
Parameters
- tnuid: tnuid
- *media_type: media_type
- *inst_id: inst_id
- *offset: Number
- *limit: Number
- format: String
- key: String
- version: Number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getTaxonMedia?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Taxon Data Type Glossary
Classes
TaxonAssociation
- assoc_taxon: TaxonRelationship
- assoc_type: assoc_type
- assoc_type_full: assoc_type_full
- assoc_occurrences: array of OccurrenceBasic
TaxonBasic
- tnuid: tnuid
- guid: guid
- tcid: tcid
- taxon: taxon
- taxon_author: taxon_author
TaxonComplete (extends TaxonRelationship)
- stats: TaxonInfoStats
- contribs: array of Contribution
- orig_desc: LiteratureExtended
TaxonExtended (extends TaxonBasic)
- describer: describer
- status: status
- valid: valid
- rank: rank
- valid_taxon: TaxonBasic
- fossil: fossil
- homonym: homonym
- common_names: array of common_name
- source: TaxonSource
TaxonHierarchy
Aggregate of one or more of the following classes:
- TaxonHierKingdom (1)
- TaxonHierPhylum (0 or 1)
- TaxonHierDivision (0 or 1)
- TaxonHierSubphylum (0 or 1)
- TaxonHierClass (0 or 1)
- TaxonHierSubclass (0 or 1)
- TaxonHierInfraclass (0 or 1)
- TaxonHierOrder (0 or 1)
- TaxonHierSuborder (0 or 1)
- TaxonHierInfraorder (0 or 1)
- TaxonHierCohort (0 or 1)
- TaxonHierSubcohort (0 or 1)
- TaxonHierSuperfamily (0 or 1)
- TaxonHierFamily (0 or 1)
- TaxonHierSubfamily (0 or 1)
- TaxonHierTribe (0 or 1)
- TaxonHierSubtribe (0 or 1)
- TaxonHierGenus (0 or 1)
- TaxonHierSpecies (0 or 1)
- TaxonHierSubspecies (0 or 1)
TaxonHierClass
- Class: TaxonHierUnit
TaxonHierCohort
- Cohort: TaxonHierUnit
TaxonHierDivision
- Division: TaxonHierUnit
TaxonHierFamily
- Family: TaxonHierUnit
TaxonHierGenus
- Genus: TaxonHierUnit
TaxonHierInfraclass
- Infraclass: TaxonHierUnit
TaxonHierInfraorder
- Infraorder: TaxonHierUnit
TaxonHierKingdom
- Kingdom: TaxonHierUnit
TaxonHierOrder
- Order: TaxonHierUnit
TaxonHierPhylum
- Phylum: TaxonHierUnit
TaxonHierSpecies
- Species: TaxonHierUnit
TaxonHierSubclass
- Subclass: TaxonHierUnit
TaxonHierSubcohort
- Subcohort: TaxonHierUnit
TaxonHierSubfamily
- Subfamily: TaxonHierUnit
TaxonHierSuborder
- Suborder: TaxonHierUnit
TaxonHierSubphylum
- Subphylum: TaxonHierUnit
TaxonHierSubspecies
- Subspecies: TaxonHierUnit
TaxonHierSubtribe
- Subtribe: TaxonHierUnit
TaxonHierSuperfamily
- Superfamily: TaxonHierUnit
TaxonHierTribe
- Tribe: TaxonHierUnit
TaxonHierUnit (extends TaxonBasic)
- next: rank
TaxonInfoStats (extends TaxonStats)
- child_stats: array of TaxonStats
- num_records: Number
- num_spms: Number
TaxonRelationship (extends TaxonExtended)
- rel_type: rel_type
- parent_taxon: TaxonBasic
- hier: TaxonHierarchy
TaxonReturn
- general: TaxonBasic
TaxonReturnAssociations (extends TaxonReturn)
- associations: array of TaxonAssociation
TaxonReturnDeterminers (extends TaxonReturn)
- determiners: array of AgentExtended
TaxonReturnHabitats (extends TaxonReturn)
- hatitats: array of habitat
TaxonReturnIncluded (extends TaxonReturn)
- includedTaxa: array of TaxonExtended
TaxonReturnInstitutions (extends TaxonReturn)
- institutions: array of InstitutionBasic
TaxonReturnLiteratureCitations (extends TaxonReturn)
- pub_citations: array of LiteratureTaxonCitationExtended
TaxonReturnLocalities (extends TaxonReturn)
- localities: array of LocalityExtended
TaxonReturnMedia (extends TaxonReturn)
- media: array of MediaExtended
TaxonReturnOccurrences (extends TaxonReturn)
- occurrences: array of OccurrenceExtended
TaxonReturnSynonyms (extends TaxonReturn)
- synonyms: array of TaxonRelationship
TaxonSource
- id: Number
- name: String
- logo: URL
- url: URL
- query: URL
TaxonStats
- rank: rank
- rank_value: rank_value
- num_taxa: Number
Elements
assoc_type
String - A description of the biological relationship between a specified taxon and another organism.
assoc_type_full
String - A complete description of the biological relationship between a specified taxon and another organism.
common_name
String - A common name for a taxon that can be included as a taxon name string but is never valid.
describer
String - the describer(s) of a taxon.
fossil
String - A modified boolean_flag, either Y, N or B, representing whether a taxon name is a extinct fossil, exclusively extant, or present today and in fossils.
guid
String - A globally unique identifier for an xBio:D domain resource in the form of a stable URI dereferenced by the bioguid.osu.edu domain.
habitat
String - Description or identifier of the ecological or environmental area that a particular species is known to inhabit.
homonym
Boolean_flag - Boolean_flag indicating whether or not the determination was a homonym
rank
String - The taxonomic (hierarchical) rank of a taxon name.
rank_value
Number - A number representing the rank of a taxon with higher numbers representing higher positions in the taxonomic hierarchy.
rel_type
String - The taxonomic (hierarchical) relationship between the taxonomic concept indicated by the taxon name and its parent taxonomic concept. Can be Member, Junior synonym, etc.
status
String - The nomenclatural status of a taxon name using a formalized vocabulary.
taxon
String - A taxon name string.
taxon_author
String - The author(s) of a taxon with parenthesis surrounding the name of the author(s) if necessary.
tcid
Number - The taxonomic concept identifier which uniquely identifies a taxon concept, which may have many name uses, within xBio:D.
tnuid
Number - The taxon name use identifier which uniquely identifies a taxon within xBio:D.
valid
String - A Valid or Invalid string representing the validity of a taxon name using subjective interpretations if warranted.
See also
Visit any of the below links to find information about the other data domains defined by OJ_Break Version 2.