Difference between revisions of "OJ Break v2 Occurrence Reference"
(→See also) |
(→API Information and Access) |
||
| (12 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
'''Introduction''' | '''Introduction''' | ||
| − | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with | + | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication. |
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | ||
| Line 10: | Line 10: | ||
Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return ''format'', an API access ''key'', and a ''version'' number. | Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return ''format'', an API access ''key'', and a ''version'' number. | ||
| − | Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format= | + | Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | ||
| + | |||
| + | |||
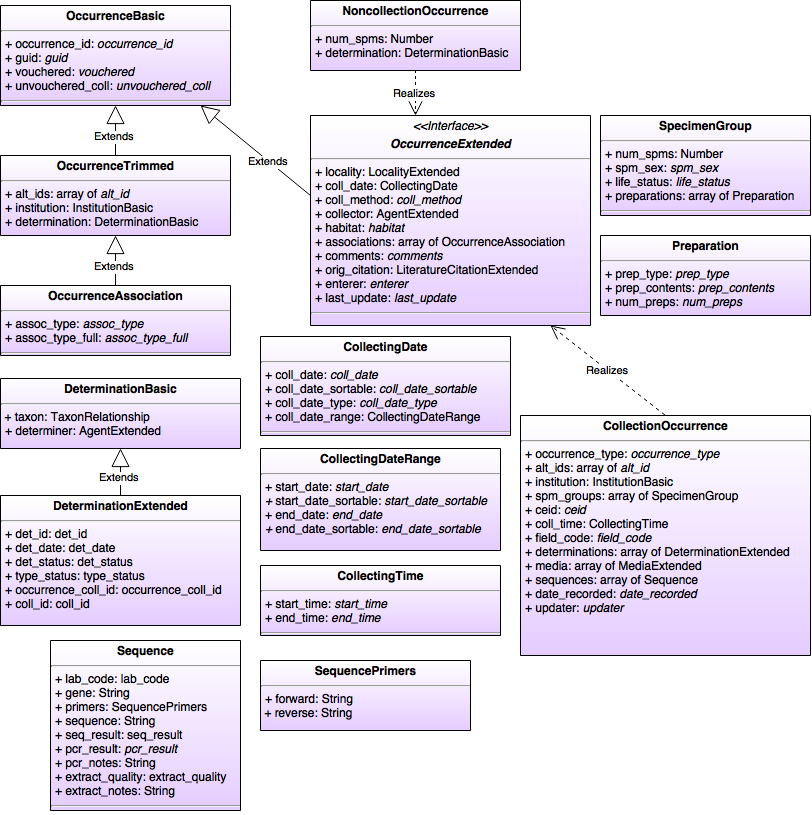
| + | [[File:OJ_Break Data Model - Occurrence.png|none|frame|Occurrence Data Model]] | ||
== Procedural Reference == | == Procedural Reference == | ||
| Line 24: | Line 27: | ||
* ''version'': number | * ''version'': number | ||
===== Return ===== | ===== Return ===== | ||
| − | * [[ | + | * [[#OccurrenceExtended (extends OccurrenceBasic)|OccurrenceExtended]] |
===== Example ===== | ===== Example ===== | ||
| − | http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format= | + | http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
==== getOccurrencesInfo ==== | ==== getOccurrencesInfo ==== | ||
| Line 37: | Line 40: | ||
* ''version'': number | * ''version'': number | ||
===== Return ===== | ===== Return ===== | ||
| − | * array of [[ | + | * array of [[#OccurrenceExtended (extends OccurrenceBasic)|OccurrenceExtended]] |
===== Example ===== | ===== Example ===== | ||
| − | http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrencesInfo?occurrence_ids=1,2&format= | + | http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrencesInfo?occurrence_ids=1,2&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
== Occurrence Data Type Glossary == | == Occurrence Data Type Glossary == | ||
| Line 57: | Line 60: | ||
* ''start_time'': ''[[#start_time|start_time]]'' | * ''start_time'': ''[[#start_time|start_time]]'' | ||
* ''end_time'': ''[[#end_time|end_time]]'' | * ''end_time'': ''[[#end_time|end_time]]'' | ||
| − | ==== CollectionOccurrence (realizes [[ | + | ==== CollectionOccurrence (realizes [[#OccurrenceExtended (extends OccurrenceBasic)|OccurrenceExtended]]) ==== |
* ''occurrence_type'': ''[[#occurrence_type|occurrence_type]]'' | * ''occurrence_type'': ''[[#occurrence_type|occurrence_type]]'' | ||
* ''alt_ids'': array of ''[[#alt_id|alt_id]]'' | * ''alt_ids'': array of ''[[#alt_id|alt_id]]'' | ||
| Line 65: | Line 68: | ||
* ''coll_time'': ''[[#CollectingTime|CollectingTime]] | * ''coll_time'': ''[[#CollectingTime|CollectingTime]] | ||
* ''field_code'': ''[[#field_code|field_code]]'' | * ''field_code'': ''[[#field_code|field_code]]'' | ||
| − | * ''determinations'': array of [[# | + | * ''determinations'': array of [[#DeterminationExtended (extends DeterminationBasic)|DeterminationExtended]] |
* ''media'': array of [[#MediaExtended|MediaExtended]] | * ''media'': array of [[#MediaExtended|MediaExtended]] | ||
* ''sequences'': array of [[#Sequence|Sequence]] | * ''sequences'': array of [[#Sequence|Sequence]] | ||
| Line 72: | Line 75: | ||
==== DeterminationBasic ==== | ==== DeterminationBasic ==== | ||
| − | * ''taxon'': [[OJ_Break v2 Taxon Reference#TaxonRelationship|TaxonRelationship]] | + | * ''taxon'': [[OJ_Break v2 Taxon Reference#TaxonRelationship (extends TaxonExtended)|TaxonRelationship]] |
| − | * ''determiner'': [[#OJ_Break v2 Agent Reference# | + | * ''determiner'': [[#OJ_Break v2 Agent Reference#AgentExtended (extends_AgentBasic)|AgentExtended]] |
| − | ==== DeterminationExtended ==== | + | |
| + | ==== DeterminationExtended (extends [[#DeterminationBasic|DeterminationBasic]]) ==== | ||
* ''det_id'': ''[[#det_id|det_id]]'' | * ''det_id'': ''[[#det_id|det_id]]'' | ||
* ''det_date'': ''[[#det_date|det_date]]'' | * ''det_date'': ''[[#det_date|det_date]]'' | ||
| Line 81: | Line 85: | ||
* ''occurrence_coll_id'': ''[[#occurrence_coll_id|occurrence_coll_id]]'' | * ''occurrence_coll_id'': ''[[#occurrence_coll_id|occurrence_coll_id]]'' | ||
* ''coll_id'': ''[[#coll_id|coll_id]]'' | * ''coll_id'': ''[[#coll_id|coll_id]]'' | ||
| − | ==== NoncollectionOccurrence ==== | + | ==== NoncollectionOccurrence (realizes [[#OccurrenceExtended (extends OccurrenceBasic)|OccurrenceExtended]]) ==== |
* ''num_spms'': Number | * ''num_spms'': Number | ||
* ''determination'': [[#DeterminationBasic|DeterminationBasic]] | * ''determination'': [[#DeterminationBasic|DeterminationBasic]] | ||
| − | ==== OccurrenceAssociation ==== | + | ==== OccurrenceAssociation (extends [[#OccurrenceTrimmed (extends OccurrenceBasic)|OccurrenceTrimmed]]) ==== |
* ''assoc_type'': ''[[#assoc_type|assoc_type]]'' | * ''assoc_type'': ''[[#assoc_type|assoc_type]]'' | ||
* ''assoc_type_full'': ''[[#assoc_type_full|assoc_type_full]]'' | * ''assoc_type_full'': ''[[#assoc_type_full|assoc_type_full]]'' | ||
| Line 93: | Line 97: | ||
* ''vouchered'': ''[[#vouchered|vouchered]]'' | * ''vouchered'': ''[[#vouchered|vouchered]]'' | ||
* ''unvouchered'': ''[[#unvouchered|unvouchered]]'' | * ''unvouchered'': ''[[#unvouchered|unvouchered]]'' | ||
| − | ==== OccurrenceExtended ==== | + | ==== OccurrenceExtended (extends [[#OccurrenceBasic|OccurrenceBasic]]) ==== |
| − | * ''locality'': [[OJ_Break v2 Locality Reference# | + | * ''locality'': [[OJ_Break v2 Locality Reference#LocalityExtended (extends LocalityBasic)|LocalityExtended]] |
* ''coll_date'': [[#CollectingDate|CollectingDate]] | * ''coll_date'': [[#CollectingDate|CollectingDate]] | ||
* ''coll_method'': [[#coll_method|coll_method]] | * ''coll_method'': [[#coll_method|coll_method]] | ||
| − | * ''collector'': [[OJ_Break v2 Agent Reference# | + | * ''collector'': [[OJ_Break v2 Agent Reference#AgentExtended (extends AgentBasic)|AgentExtended]] |
* ''habitat'': ''[[#habitat|habitat]]'' | * ''habitat'': ''[[#habitat|habitat]]'' | ||
* ''associations'': array of [[#OccurrenceAssociation|OccurrenceAssociation]] | * ''associations'': array of [[#OccurrenceAssociation|OccurrenceAssociation]] | ||
* ''comments'': ''[[#comments|comments]]'' | * ''comments'': ''[[#comments|comments]]'' | ||
| − | * ''orig_citation'': [[OJ_Break v2 | + | * ''orig_citation'': [[OJ_Break v2 Literature Reference#LiteratureCitationExtended (extends LiteratureCitationBasic)|LiteratureCitationExtended]] |
* ''enterer'': ''[[#enterer|enterer]]'' | * ''enterer'': ''[[#enterer|enterer]]'' | ||
* ''last_update'': ''[[#last_update|last_update]]'' | * ''last_update'': ''[[#last_update|last_update]]'' | ||
| − | ==== OccurrenceTrimmed ==== | + | ==== OccurrenceTrimmed (extends [[#OccurrenceBasic|OccurrenceBasic]]) ==== |
* ''alt_ids'': array of ''alt_id'' | * ''alt_ids'': array of ''alt_id'' | ||
| − | * ''institution'': [[InstitutionBasic]] | + | * ''institution'': [[OJ_Break v2 Institution Reference#InstitutionBasic|InstitutionBasic]] |
* ''determination'': [[#DeterminationBasic|DeterminationBasic]] | * ''determination'': [[#DeterminationBasic|DeterminationBasic]] | ||
| + | |||
==== Preparation ==== | ==== Preparation ==== | ||
* ''prep_type'': ''[[#prep_type|prep_type]]'' | * ''prep_type'': ''[[#prep_type|prep_type]]'' | ||
| Line 132: | Line 137: | ||
=== Elements === | === Elements === | ||
| + | ==== alt_id ==== | ||
| + | String - An alternate identifier for the specimen. | ||
| + | ==== assoc_type ==== | ||
| + | String - A description of the biological relationship between a specified taxon and another organism. | ||
| + | ==== assoc_type_full ==== | ||
| + | String - A complete description of the biological relationship between a specified taxon and another organism. | ||
| + | ==== ceid ==== | ||
| + | Number - | ||
| + | ==== coll_date ==== | ||
| + | String - | ||
| + | ==== coll_date_sortable ==== | ||
| + | String - | ||
| + | ==== coll_id ==== | ||
| + | Number - | ||
| + | ==== coll_method ==== | ||
| + | String | ||
| + | ==== comments ==== | ||
| + | String - | ||
| + | ==== date_recorded ==== | ||
| + | String | ||
| + | ==== det_date ==== | ||
| + | String | ||
| + | ==== det_id ==== | ||
| + | Number | ||
| + | ==== det_status ==== | ||
| + | String | ||
| + | ==== end_date ==== | ||
| + | String | ||
| + | ==== end_date_sortable ==== | ||
| + | String | ||
| + | ==== end_time ==== | ||
| + | String | ||
| + | ==== enterer ==== | ||
| + | String | ||
| + | ==== extract_quality ==== | ||
| + | String | ||
| + | ==== field_code ==== | ||
| + | String | ||
| + | ==== guid ==== | ||
| + | GUID - Globally Unique Identifier that uniquely identifies a resource. | ||
| + | ==== habitat ==== | ||
| + | String | ||
| + | ==== lab_code ==== | ||
| + | String | ||
| + | ==== last_update ==== | ||
| + | String | ||
| + | ==== life_status ==== | ||
| + | String | ||
| + | ==== num_preps ==== | ||
| + | Number - Number of preparations. | ||
| + | ==== occurrence_coll_id ==== | ||
| + | Number | ||
| + | ==== occurrence_id ==== | ||
| + | Number | ||
| + | ==== occurrence_type ==== | ||
| + | String | ||
| + | ==== seq_result ==== | ||
| + | String | ||
| + | ==== spm_sex ==== | ||
| + | String | ||
| + | ==== start_time ==== | ||
| + | String | ||
| + | ==== pcr_result ==== | ||
| + | String | ||
| + | ==== prep_contents ==== | ||
| + | String | ||
| + | ==== prep_type ==== | ||
| + | String | ||
| + | ==== type_status ==== | ||
| + | String | ||
| + | ==== unvouchered_coll ==== | ||
| + | ? | ||
| + | ==== updater ==== | ||
| + | String | ||
| + | ==== vouchered ==== | ||
| + | Boolean_flag | ||
| + | |||
== See also == | == See also == | ||
Visit any of the below links to find information about the other data domains defined by OJ_Break Version 2. | Visit any of the below links to find information about the other data domains defined by OJ_Break Version 2. | ||
Latest revision as of 16:39, 9 June 2015
Introduction
OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication.
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database.
Contents
- 1 API Information and Access
- 2 Procedural Reference
- 3 Occurrence Data Type Glossary
- 3.1 Classes
- 3.1.1 CollectingDate
- 3.1.2 CollectingDateRange
- 3.1.3 CollectingTime
- 3.1.4 CollectionOccurrence (realizes OccurrenceExtended)
- 3.1.5 DeterminationBasic
- 3.1.6 DeterminationExtended (extends DeterminationBasic)
- 3.1.7 NoncollectionOccurrence (realizes OccurrenceExtended)
- 3.1.8 OccurrenceAssociation (extends OccurrenceTrimmed)
- 3.1.9 OccurrenceBasic
- 3.1.10 OccurrenceExtended (extends OccurrenceBasic)
- 3.1.11 OccurrenceTrimmed (extends OccurrenceBasic)
- 3.1.12 Preparation
- 3.1.13 Sequence
- 3.1.14 SequencePrimers
- 3.1.15 SpecimenGroup
- 3.2 Elements
- 3.2.1 alt_id
- 3.2.2 assoc_type
- 3.2.3 assoc_type_full
- 3.2.4 ceid
- 3.2.5 coll_date
- 3.2.6 coll_date_sortable
- 3.2.7 coll_id
- 3.2.8 coll_method
- 3.2.9 comments
- 3.2.10 date_recorded
- 3.2.11 det_date
- 3.2.12 det_id
- 3.2.13 det_status
- 3.2.14 end_date
- 3.2.15 end_date_sortable
- 3.2.16 end_time
- 3.2.17 enterer
- 3.2.18 extract_quality
- 3.2.19 field_code
- 3.2.20 guid
- 3.2.21 habitat
- 3.2.22 lab_code
- 3.2.23 last_update
- 3.2.24 life_status
- 3.2.25 num_preps
- 3.2.26 occurrence_coll_id
- 3.2.27 occurrence_id
- 3.2.28 occurrence_type
- 3.2.29 seq_result
- 3.2.30 spm_sex
- 3.2.31 start_time
- 3.2.32 pcr_result
- 3.2.33 prep_contents
- 3.2.34 prep_type
- 3.2.35 type_status
- 3.2.36 unvouchered_coll
- 3.2.37 updater
- 3.2.38 vouchered
- 3.1 Classes
- 4 See also
API Information and Access
This page specifies the methods and data defined by OJ_Break Version 2, more precisely those that are defined by the Occurrence data domain. To get information on any of the other data domains defined in OJ_Break Version 2, visit the See also section.
Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return format, an API access key, and a version number.
Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Notice the specification of the version parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the version parameter has a default value of 2 making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to OJ_Break API Access.
Procedural Reference
Occurrence
getOccurrenceInfo
Description
Parameters
- occurrence_id: occurrence_id
- format: string
- key: string
- version: number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrenceInfo?occurrence_id=1&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getOccurrencesInfo
Description
Parameters
- occurrence_ids: array of occurrence_id
- format: string
- key: string
- version: number
Return
- array of OccurrenceExtended
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getOccurrencesInfo?occurrence_ids=1,2&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Occurrence Data Type Glossary
Classes
CollectingDate
- coll_date: coll_date
- coll_date_sortable: coll_date_sortable
- coll_date_type: coll_date_type
- coll_date_range: CollectingDateRange
CollectingDateRange
- start_date: start_date
- start_date_sortable: start_date_sortable
- end_date: end_date
- end_date_sortable: end_date_sortable
CollectingTime
- start_time: start_time
- end_time: end_time
CollectionOccurrence (realizes OccurrenceExtended)
- occurrence_type: occurrence_type
- alt_ids: array of alt_id
- institution: InstitutionBasic
- spm_groups: array of SpecimenGroup
- ceid: ceid
- coll_time: CollectingTime
- field_code: field_code
- determinations: array of DeterminationExtended
- media: array of MediaExtended
- sequences: array of Sequence
- date_recorded: date_recorded
- updater: updater
DeterminationBasic
- taxon: TaxonRelationship
- determiner: AgentExtended
DeterminationExtended (extends DeterminationBasic)
- det_id: det_id
- det_date: det_date
- det_status: det_status
- type_status: type_status
- occurrence_coll_id: occurrence_coll_id
- coll_id: coll_id
NoncollectionOccurrence (realizes OccurrenceExtended)
- num_spms: Number
- determination: DeterminationBasic
OccurrenceAssociation (extends OccurrenceTrimmed)
- assoc_type: assoc_type
- assoc_type_full: assoc_type_full
OccurrenceBasic
- occurrence_id: occurrence_id
- guid: guid
- vouchered: vouchered
- unvouchered: unvouchered
OccurrenceExtended (extends OccurrenceBasic)
- locality: LocalityExtended
- coll_date: CollectingDate
- coll_method: coll_method
- collector: AgentExtended
- habitat: habitat
- associations: array of OccurrenceAssociation
- comments: comments
- orig_citation: LiteratureCitationExtended
- enterer: enterer
- last_update: last_update
OccurrenceTrimmed (extends OccurrenceBasic)
- alt_ids: array of alt_id
- institution: InstitutionBasic
- determination: DeterminationBasic
Preparation
- prep_type: prep_type
- prep_contents: prep_contents
- num_preps: num_preps
Sequence
- lab_code: lab_code
- gene: String
- primers: SequencePrimers
- sequence: String
- seq_result: seq_result
- pcr_result: pcr_result
- pcr_notes: String
- extract_quality: extract_quality
- extract_notes: String
SequencePrimers
- forward: String
- reverse: String
SpecimenGroup
- num_spms: Number
- spm_sex: spm_sex
- life_status: life_status
- preparations: array of Preparation
Elements
alt_id
String - An alternate identifier for the specimen.
assoc_type
String - A description of the biological relationship between a specified taxon and another organism.
assoc_type_full
String - A complete description of the biological relationship between a specified taxon and another organism.
ceid
Number -
coll_date
String -
coll_date_sortable
String -
coll_id
Number -
coll_method
String
comments
String -
date_recorded
String
det_date
String
det_id
Number
det_status
String
end_date
String
end_date_sortable
String
end_time
String
enterer
String
extract_quality
String
field_code
String
guid
GUID - Globally Unique Identifier that uniquely identifies a resource.
habitat
String
lab_code
String
last_update
String
life_status
String
num_preps
Number - Number of preparations.
occurrence_coll_id
Number
occurrence_id
Number
occurrence_type
String
seq_result
String
spm_sex
String
start_time
String
pcr_result
String
prep_contents
String
prep_type
String
type_status
String
unvouchered_coll
?
updater
String
vouchered
Boolean_flag
See also
Visit any of the below links to find information about the other data domains defined by OJ_Break Version 2.