Difference between revisions of "OJ Break v2 Journal Reference"
(→JournalReturnTaxa (JournalReturn)) |
(→API Information and Access) |
||
| (5 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
'''Introduction''' | '''Introduction''' | ||
| − | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with | + | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication. |
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | ||
| Line 10: | Line 10: | ||
Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return ''format'', an API access ''key'', and a ''version'' number. | Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return ''format'', an API access ''key'', and a ''version'' number. | ||
| − | Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format= | + | Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | ||
| + | |||
| + | |||
| + | [[File:OJ_Break Data Model - Journal.png|none|frame|Journal Data Model]] | ||
== Procedural Reference == | == Procedural Reference == | ||
| Line 27: | Line 30: | ||
===== Example ===== | ===== Example ===== | ||
| − | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format= | + | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
==== getJournalLiterature ==== | ==== getJournalLiterature ==== | ||
| Line 39: | Line 42: | ||
* ''version'': number | * ''version'': number | ||
===== Return ===== | ===== Return ===== | ||
| − | * [[# | + | * [[#JournalReturnLiterature (extends JournalReturn)|JournalReturnLiterature]] |
===== Example ===== | ===== Example ===== | ||
| − | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalLiterature?journal_id=33&format= | + | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalLiterature?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
==== getJournalTaxa ==== | ==== getJournalTaxa ==== | ||
| Line 54: | Line 57: | ||
* ''version'': number | * ''version'': number | ||
===== Return ===== | ===== Return ===== | ||
| − | * [[# | + | * [[#JournalReturnTaxa (extends JournalReturn)|JournalReturnTaxa]] |
===== Example ===== | ===== Example ===== | ||
| − | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalTaxa?journal_id=33&format= | + | http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalTaxa?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
== Journal Data Type Glossary == | == Journal Data Type Glossary == | ||
| Line 66: | Line 69: | ||
* ''journal_abbrev'': [[#journal_abbrev|journal_abbrev]] | * ''journal_abbrev'': [[#journal_abbrev|journal_abbrev]] | ||
| − | ==== JournalComplete (extends [[# | + | ==== JournalComplete (extends [[#JournalExtended (extends JournalBasic)|JournalExtended]]) ==== |
* ''stats'': [[#JournalStats|JournalStats]] | * ''stats'': [[#JournalStats|JournalStats]] | ||
| Line 78: | Line 81: | ||
==== JournalReturnLiterature (extends [[#JournalReturn|JournalReturn]]) ==== | ==== JournalReturnLiterature (extends [[#JournalReturn|JournalReturn]]) ==== | ||
| − | * ''literature'': array of [[#OJ_Break v2 Literature Reference# | + | * ''literature'': array of [[#OJ_Break v2 Literature Reference#LiteratureExtended (extends LiteratureBasic)|LiteratureExtended]] |
==== JournalReturnTaxa (extends [[#JournalReturn|JournalReturn]]) ==== | ==== JournalReturnTaxa (extends [[#JournalReturn|JournalReturn]]) ==== | ||
| − | * ''taxa'': array of [[OJ_Break v2 Taxon Reference# | + | * ''taxa'': array of [[OJ_Break v2 Taxon Reference#TaxonRelationship (extends TaxonExtended)|TaxonRelationship]] |
==== JournalStats ==== | ==== JournalStats ==== | ||
| Line 91: | Line 94: | ||
=== Elements === | === Elements === | ||
==== journal_id ==== | ==== journal_id ==== | ||
| − | Number - | + | Number - A numeric journal identifier that uniquely identifies a journal. |
==== journal_name ==== | ==== journal_name ==== | ||
| − | String - | + | String - A journal name string. |
==== journal_abbrev ==== | ==== journal_abbrev ==== | ||
| − | String - | + | String - A abbreviated journal name string. |
==== journal_url ==== | ==== journal_url ==== | ||
| − | URL - | + | URL - A URL to a journal. |
==== journal_copyright ==== | ==== journal_copyright ==== | ||
| − | String - | + | String - Copyright details for a journal. |
==== public ==== | ==== public ==== | ||
| − | Boolean_flag - | + | Boolean_flag - A Y or N indicator that signifies whether a journal is made public or not. |
== See also == | == See also == | ||
| Line 111: | Line 114: | ||
* [[OJ_Break v2 Literature Reference|Literature]] | * [[OJ_Break v2 Literature Reference|Literature]] | ||
* [[OJ_Break v2 Locality Reference|Locality]] | * [[OJ_Break v2 Locality Reference|Locality]] | ||
| + | * [[OJ_Break v2 Media Reference|Media]] | ||
* [[OJ_Break v2 Occurrence Reference|Occurrence]] | * [[OJ_Break v2 Occurrence Reference|Occurrence]] | ||
* [[OJ_Break v2 Search Reference|Search]] | * [[OJ_Break v2 Search Reference|Search]] | ||
* [[OJ_Break v2 Taxon Reference|Taxon]] | * [[OJ_Break v2 Taxon Reference|Taxon]] | ||
Latest revision as of 16:36, 9 June 2015
Introduction
OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with HTML, JSON, XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication.
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database.
Contents
API Information and Access
This page specifies the methods and data defined by OJ_Break Version 2, more precisely those that are defined by the Journal data domain. To get information on any of the other data domains defined in OJ_Break Version 2, visit the See also section.
Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found on this page) and a few other required parameters. These required parameters include specifying a return format, an API access key, and a version number.
Example: http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Notice the specification of the version parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the version parameter has a default value of 2 making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to OJ_Break API Access.
Procedural Reference
Journal
getJournalInfo
Description
Parameters
- journal_id: journal_id
- format: string
- key: string
- version: number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalInfo?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getJournalLiterature
Description
Parameters
- journal_id: journal_id
- *offset: number
- *limit: number
- format: string
- key: string
- version: number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalLiterature?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
getJournalTaxa
Description
Parameters
- journal_id: journal_id
- *offset: number
- *limit: number
- format: string
- key: string
- version: number
Return
Example
http://osuc.biosci.ohio-state.edu/OJ_Break/getJournalTaxa?journal_id=33&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Journal Data Type Glossary
Classes
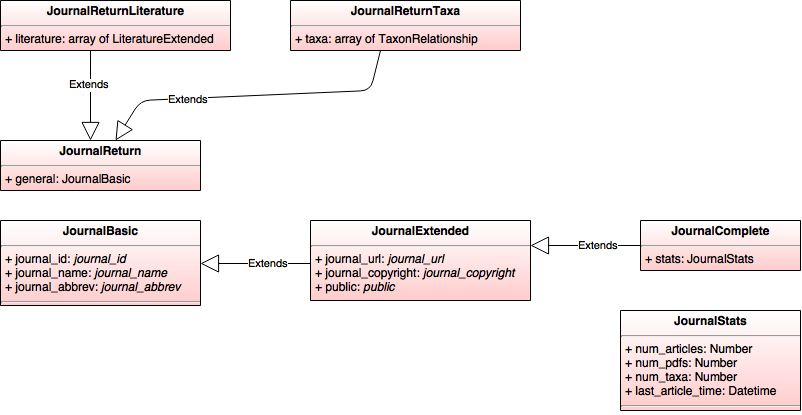
JournalBasic
- journal_id: journal_id
- journal_name: journal_name
- journal_abbrev: journal_abbrev
JournalComplete (extends JournalExtended)
- stats: JournalStats
JournalExtended (extends JournalBasic)
- journal_url: journal_url
- journal_copyright: journal_copyright
- public: public
JournalReturn
- general: JournalBasic
JournalReturnLiterature (extends JournalReturn)
- literature: array of LiteratureExtended
JournalReturnTaxa (extends JournalReturn)
- taxa: array of TaxonRelationship
JournalStats
- num_articles: Number
- num_pdfs: Number
- num_taxa: Number
- last_article_time: Datetime
Elements
journal_id
Number - A numeric journal identifier that uniquely identifies a journal.
journal_name
String - A journal name string.
journal_abbrev
String - A abbreviated journal name string.
journal_url
URL - A URL to a journal.
journal_copyright
String - Copyright details for a journal.
public
Boolean_flag - A Y or N indicator that signifies whether a journal is made public or not.
See also
Visit any of the below links to find information about the other data domains defined by OJ_Break Version 2.