Difference between revisions of "OJ Break Version 2 API Reference"
(Updated the url to now point to the new server) |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
'''Introduction''' | '''Introduction''' | ||
| − | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with JavaScript Object Notation (JSON), XML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication. | + | OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with JavaScript Object Notation (JSON), XML, simple HTML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication. |
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database. | ||
| Line 9: | Line 9: | ||
This page specifies the methods and data defined by OJ_Break Version 2. Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found in linked pages) and a few other required parameters. These include specifying a return ''format'', an API access ''key'', and a ''version'' number. | This page specifies the methods and data defined by OJ_Break Version 2. Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found in linked pages) and a few other required parameters. These include specifying a return ''format'', an API access ''key'', and a ''version'' number. | ||
| − | Example: | + | Example: https://osuc.biosci.ohio-state.edu/cgi-bin/OJ_Break_API.py/getTaxonInfo?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2 |
Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | Notice the specification of the ''version'' parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the ''version'' parameter has a default value of ''2'' making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to [[OJ_Break API Access]]. | ||
| + | |||
| + | == API Calls == | ||
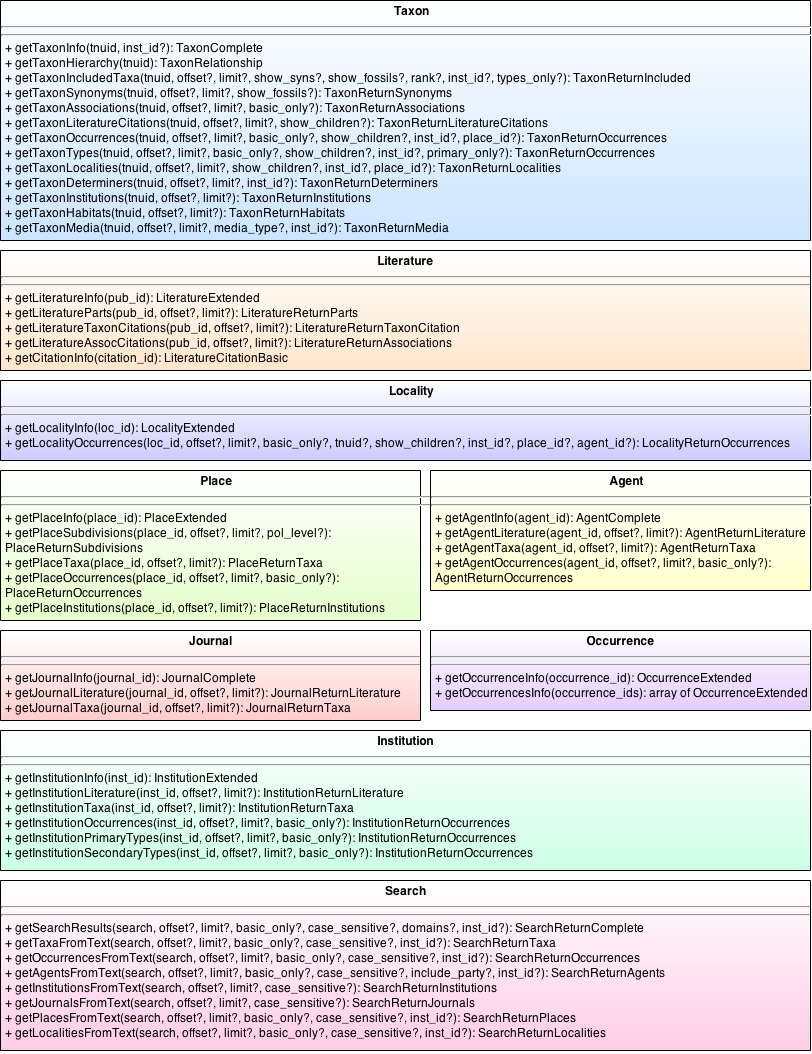
| + | The API calls for version 2 of the OJ_Break API are listed in the diagram below. The calls are organized into their respective data domains and present the available parameters and return class for each call. | ||
| + | |||
| + | [[File:OJ Break API Calls.png|none|frame|Complete Set of API Calls]] | ||
== Data Domains == | == Data Domains == | ||
| Line 21: | Line 26: | ||
* [[OJ_Break v2 Literature Reference|Literature]] | * [[OJ_Break v2 Literature Reference|Literature]] | ||
* [[OJ_Break v2 Locality Reference|Locality]] | * [[OJ_Break v2 Locality Reference|Locality]] | ||
| + | * [[OJ_Break v2 Media Reference|Media]] | ||
* [[OJ_Break v2 Occurrence Reference|Occurrence]] | * [[OJ_Break v2 Occurrence Reference|Occurrence]] | ||
* [[OJ_Break v2 Search Reference|Search]] | * [[OJ_Break v2 Search Reference|Search]] | ||
* [[OJ_Break v2 Taxon Reference|Taxon]] | * [[OJ_Break v2 Taxon Reference|Taxon]] | ||
Latest revision as of 14:59, 9 July 2018
Introduction
OJ_Break is the name of the xBio:D RESTful API service to facilitate discovery of data within the xBio:D database. The API can respond with JavaScript Object Notation (JSON), XML, simple HTML, or JSON with padding (JSONP) and accepts HTTP GET and POST requests indiscriminately. The backend of the API is written in Oracle's PL/SQL database language, which is fast but often inflexible, while a Python presentation layer mitigates request handling and authentication.
OJ_Break Version 2 is a new and improved edition of the work started in Version 1. Version 2 introduces a completely restructured and standardized data model to further enhance the functionality of the xBio:D database.
API Information and Access
This page specifies the methods and data defined by OJ_Break Version 2. Using the OJ_Break Version 2 API requires calling methods with corresponding, method specific parameters (found in linked pages) and a few other required parameters. These include specifying a return format, an API access key, and a version number.
Example: https://osuc.biosci.ohio-state.edu/cgi-bin/OJ_Break_API.py/getTaxonInfo?tnuid=30148&format=html&key=FBF57A9F7A666FC0E0430100007F0CDC&version=2
Notice the specification of the version parameter at the end of the example method call. With the introduction of OJ_Break Version 2, the version parameter has a default value of 2 making the specification in the example unnecessary. To read more about using the OJ_Break Version 2 API, go to OJ_Break API Access.
API Calls
The API calls for version 2 of the OJ_Break API are listed in the diagram below. The calls are organized into their respective data domains and present the available parameters and return class for each call.
Data Domains
As part of Version 2's effort to standardize and simplify the flow of database data, Version 2 API calls are organized into several different domains based on the data they receive and/or describe. A complete list of the data domains is given below. To see the calls described by each data domain, visit the corresponding domain page.